Data Sources¶
MEDIator version 1.0-SNAPSHOT has extensively been developed for The Cancer Imaging Archive (TCIA), while maintaining relevant interfaces for extension to the otehr data sources.
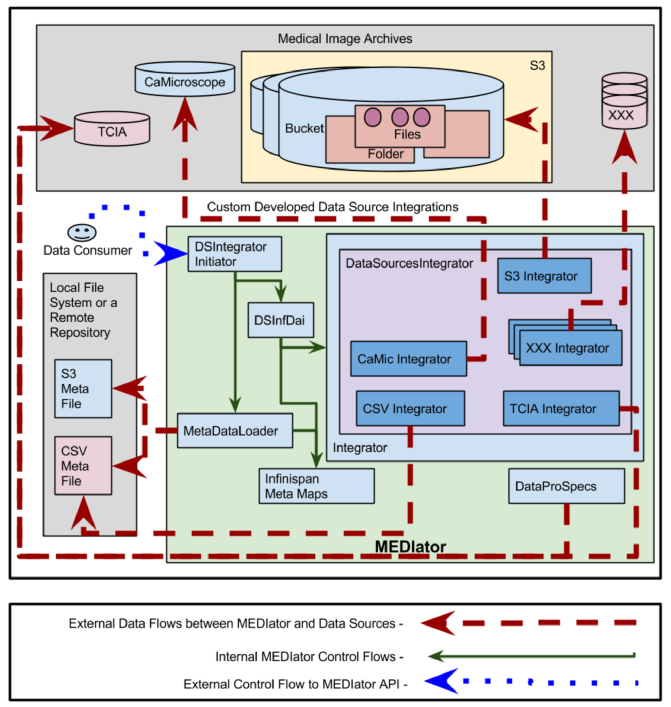
Integration with Medical Data Sources¶
Clinical data is deployed in multiple data sources such as TCIA, caMicroscope, and Amazon S3. The above figure depicts the deployment of the system with multiple medical data sources. A set of clinical data was uploaded to S3, where the metadata mapping of patientID ding{213} fileName was available as a CSV file. Similarly CSV file depicting clinical information is available, as multiple properties against the UUID, such as patient ID. These files are parsed and stored
into metadata maps in Infinispan. The CSV files containing metadata or $resource ID ding{213} fileName$ mapping are stored locally in the file system or in a remote repository.
Each data source is connected to MEDIator by implementing a class that extends Integrator interface. DatasourcesIntegrator, an abstract class provides the common features for the data sources integration. It is extended by CsvIntegrator, CaMicIntegrator, S3Integrator, and TciaIntegrator which respectively function as the integrators for CSV, caMicroscope, Amazon S3, and TCIA. MetaDataLoader loads the CSV files and stores them into Infinispan maps, against the ID, such as the patient ID.
A sub class of InfDataAccessIntegration, DSInfDai holds the instances of map to store all the metadata. DSIntegratorInitiator invokes the instances of DSInfDai and MetaDataLoader to parse the metadata, and store the instances into the respective maps.
Medical Image Archives¶
- Medical images are stored in specific data sources such as TCIA that defines a schema for the metadata, or can be stored
- in a general-purpose data source such as Amazon S3, with user-enforced schema. TCIA public API provides methods to retrieve the images and metadata of different granularity. These methods are invoked by the public APIs such as the REST API. An initial search on TCIA may contain parameters such as modality, in addition to collection name, patient ID, study instance ID, and series instance UID. Each of the searches in TCIA returns the output in a finer granularity.
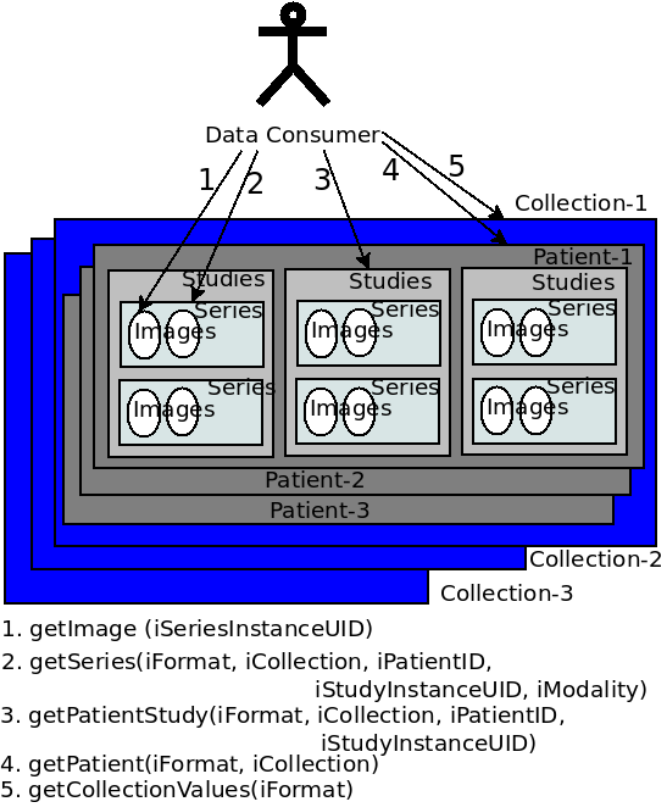
TCIA is made of multiple collections. Collections are queried and raw image data and metadata are downloaded by users. While some collections are open for public access, some are protected. Downloads create a considerable load in the servers, with unexpected spikes resulting from automated bulk downloads. The National Lung Screening Trial (NLST) 2 has a mean download rate of 0.5 TB/month, with a spike of 5.6 TB in June 2013, during the last 3 years.